Genome Editing and Perturbations with Miniature CRISPR Nucleases
This technology provides systems, methods, and compositions of miniature CRISPR-associated (Cas) nucleases to edit genes and control their activation and inhibition. This technology has applicability in the development of diagnostic tools, in addition to gene therapies.
Researchers
-
systems, methods, and compositions comprising miniature crispr nucleases for gene editing and programmable gene activation and inhibition
United States of America | Pending -
systems, methods, and compositions comprising miniature crispr nucleases for gene editing and programmable gene activation and inhibition
Canada | Published application -
systems, methods, and compositions comprising miniature crispr nucleases for gene editing and programmable gene activation and inhibition
Australia | Pending -
systems, methods, and compositions comprising miniature crispr nucleases for gene editing and programmable gene activation and inhibition
European Patent Convention | Published application -
systems, methods, and compositions comprising miniature crispr nucleases for gene editing and programmable gene activation and inhibition
Japan | Pending
Figures
Technology
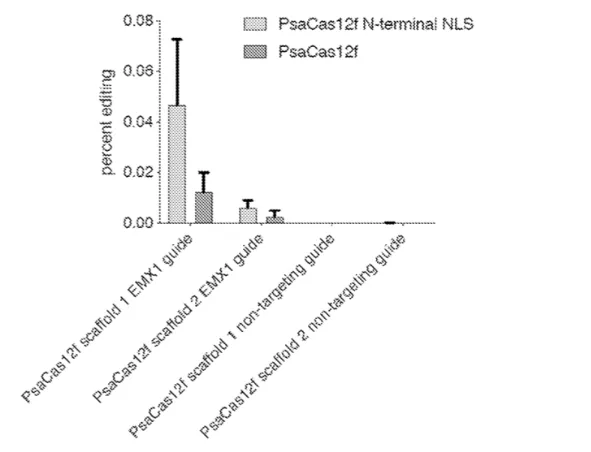
The technology is a miniature CRISPR nuclease that is target specific, compact in structure, and made up of a small number of amino acids (<800-1000). The nuclease targets DNA and is directed by a guide RNA (gRNA) to a target nucleic acid sequence, which is commonly a protospacer adjacent motif (PAM). The gRNA can be a single-guide RNA, which is a blend of two non-coding RNAs: a synthetic CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA). This system allows for the insertion or deletion of base pairs in DNA. The target specific nuclease cleaves the DNA at a target site, leaving overhangs on both ends of the DNA. Then, in nucleotide insertion, a nucleotide is added complementary to the overhanging nucleotide on both ends. In deletion, the overhanging nucleotides on both ends are removed. Finally, the DNA ends are ligated together to complete the insertion or deletion of base pairs.
Problem Addressed
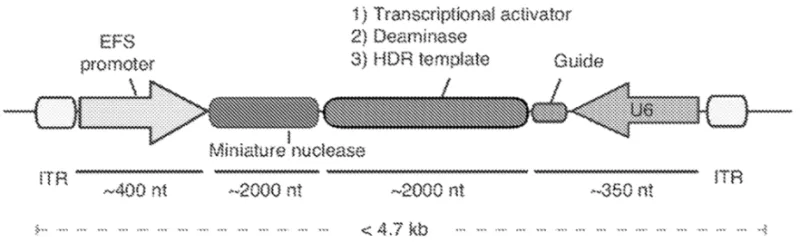
Application of CRISPR-Cas gene-editing tools without the need for repair-pathway dependent editing in mammalian cells is challenging because it requires the expression and delivery of as many as 4-7 proteins to the nucleus for proper assembly and DNA targeting. Furthermore, the ability to use Cas9 and Cas12 nucleases in CRISPR-Cas gene editing systems in vivo or ex vivo is hindered by their size, which can be as many as 1000 amino acids long. Therefore, technologies that edit or program the activation or inhibition of genes based on these nucleases cannot typically be delivered to mice by known viral vectors including the therapeutically validated adeno-associated vectors (AAV) because of their restricted size capacity (<5 kb). Smaller CRISPR-associated nucleases described here would allow for simpler delivery and extended CRISPR applications by enabling delivery of the nuclease and guide RNA in a single vector.
Advantages
- Small size allows for therapeutic applications not previously possible, including single vector homology directed repair (HDR), epigenetic editing, gene activation and repression, base editing, and prime editing
- Wide variety of potential applications provides flexibility for a licensing company
License this technology
Interested in this technology? Connect with our experienced licensing team to initiate the process.
Sign up for technology updates
Sign up now to receive the latest updates on cutting-edge technologies and innovations.